I am interested in the biogenesis, addressing and function of membrane proteins in diderm bacteria. As a PhD student at the Pasteur Institute of Paris (France), I worked on Bordetella pertussis adenylated cyclase, and its use as a genetic tool to study proteolytic enzymes. I then moved to NIH (National Institute of Health, Bethesda, USA) as a post-doctoral fellow, where I studied the Type V secretion system of Gram-negative bacteria. I continued working on this secretion system for 3 additional years as an Assistant professor at the Catholic University of America (Washington DC, USA). In 2012, I moved back to France, where I obtained a CNRS position, initially working on envelop proteins in Corynebacteriales, now investigating the mechanisms of heme import in the human pathogen Haemophilus influenzae.
Current research project
Background
Haemophilus influenzae (Hi) is an opportunistic, Gram-negative bacterium that causes a range of human infections, including otitis media, meningitis, epiglottitis and pneumonia. Although a vaccine is available since the 80’s, it does not target the non-typeable Hi strains, for which increased antibiotic resistance is a major concern. As a consequence, the World Health Organization included, in 2017, H .influenzae in the list of high-priority antimicrobial-resistant bacteria.1
Because Hi has an absolute growth requirement for heme, the systems involved in the import of this molecule represent attractive targets for new antimicrobial therapies. Such systems have indeed been successfully targeted in other microorganisms. However, the development of this type of approaches requires a precise understanding of the mechanisms involved, which is currently lacking in the case of H. influenzae.2
In the human host, the majority of heme is associated with hemoglobin in red blood cells. When hemolysis occurs, free hemoglobin (Hb) is scavenged by the high affinity serum haptoglobin (Hp) (Kd=10-15M) which targets Hb for degradation by the reticuloendothelial system. Free heme, in contrast, is detoxified after capture by hemopexin, another high affinity serum protein (Kd <pM). Hemopexin and haptoglobin hence prevent heme oxidative and pro-inflammatory damages and preclude renal toxicity caused by extracellular hemoglobin. Importantly, these proteins also participate in a process called “nutritional immunity” by restricting pathogens access to an essential nutrient (heme) and thus preventing certain infections. Indeed, hemopexin limits in vivo growth of Citrobacter rodentium and Escherichia coli, while the binding of haptoglobin to hemoglobin inhibits heme acquisition by Staphylococcus aureus and E. coli.3
H. influenzae, in contrast, is one of the very few microorganisms able to use heme-hemopexin or the hemoglobin-haptoglobin complex (Hb-Hp) as heme sources, which makes this bacterium a remarkably original model to study.
Like other Gram negative bacteria, Hi rely on outer membrane proteins belonging to the TonB Dependent Transporter (TBDT) family to import heme form the host hemoproteins.4 However, the precise molecular mechanism of heme import has not been deciphered yet.
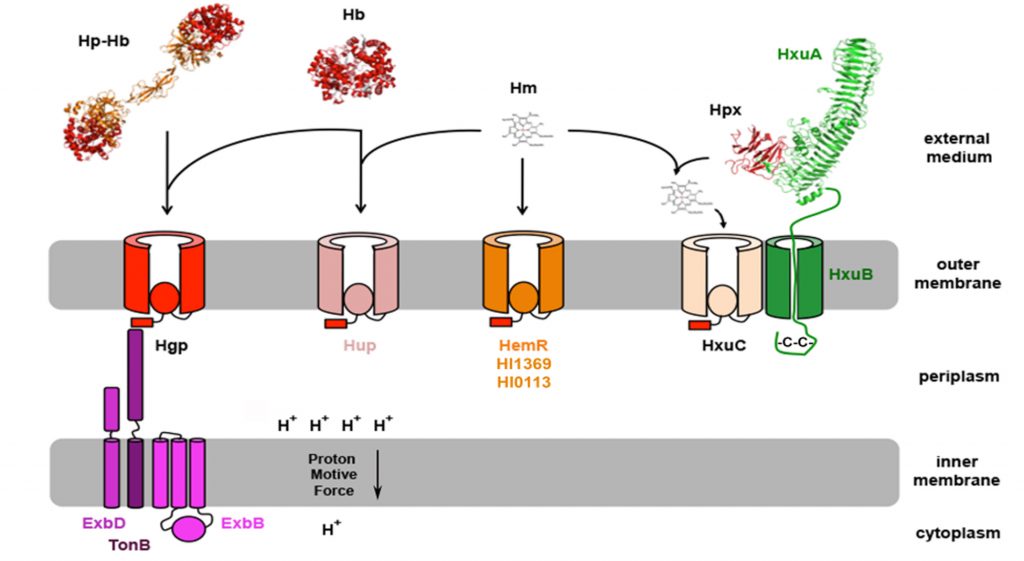
Figure 1: Heme acquisition in Haemophilus influenzae. Heme from hemoglobin (Hb) and hemoglobin-haptoglobin (Hb-Hp) can be acquired through the TBDTs Hgp and Hup. HemR could be involved in acquisition of free heme (Hm), together with HxuC and 2 uncharacterized TBDTs: HI1369 and HI0113. Heme from heme-hemopexin (Hpx) is acquired through HxuC and the acquisition is dependent on the presence of both HxuB and HxuA.
Objectives
Together with Dr. Philippe Delepelaire , I am currently working on 2 heme acquisition systems from H. influenzae: the HxuCBA system, necessary for heme acquisition from hemopexin5,6 and the Hgps, necessary for heme import form the hemoglobin-haptoglobin complex7.
Our goal is to precisely understand how these systems function. To do so, we will use a combination of in vivo and in vitro approaches (system reconstitution in E. coli, biochemical characterization of purified proteins), as well as structural characterization by cryo-EM (collaboration with Dr. Valerie Biou) and X-ray crystallography (collaboration Dr. Vincent Villeret, Lille).
References
WHO Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. 2017 www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf
Shisaka Y, Iwai Y, Yamada S, Uehara H, Tosha T, Sugimoto H, Shiro Y, Stanfield JK, Ogawa K, Watanabe Y, Shoji O. 2019. Hijacking the heme acquisition system of Pseudomonas aeruginosa for the delivery of phthalocyanine as an antimicrobial. ACS Chem. Biol. 14: 1634-1642.
Smith A, McCulloh RJ. 2015. Hemopexin and haptoglobin: allies against heme toxicity from hemoglobin not contenders. Front Physiol. 6:187.
Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Rev. Microbiol. 64:43-60.
Fournier C, Smith A, Delepelaire P. 2011. Haem release from haemopexin by HxuA allows Haemophilus influenzae to escape host nutritional immunity. Microbiol. 80: 133-48.
Zambolin S, Clantin B, Chami M, Hoos S, Haouz A, Villeret V, Delepelaire P. 2016. Structural basis for haem piracy from host haemopexin by Haemophilus influenzae. Commun. 7: 11590.
Morton DJ, Whitby PW, Jin H, Ren Z, Stull TL. 1999. Effect of multiple mutations in the hemoglobin- and hemoglobin-haptoglobin-binding proteins, HgpA, HgpB, and HgpC, of Haemophilus influenzae type b. Immun. 67:2729-39.