Bruno
Miroux
Research Director
Francesca
Zito
Research Director

Mukul
Kareya
Post-doc

Céline
Madigou
Enginneer

Dhenesh
Puvanendran
UPC MCU
Membrane biogenesis in E. coli
Context:
Membrane remodeling and phospholipid biosynthesis are normally tightly regulated to maintain the shape and function of cells. Indeed, different physiological mechanisms ensure a precise coordination between de novo phospholipid biosynthesis and modulation of membrane morphology. Interestingly, the overproduction of certain membrane proteins hijack these regulation networks, leading to the formation of impressive intracellular membrane structures in both prokaryotic and eukaryotic cells (Figure 1). The proteins triggering an abnormal accumulation of membrane structures inside the cells (or membrane proliferation) share two major common features: i, they promote the formation of highly curved membrane domains and ii, they lead to an enrichment in anionic, cone-shaped phospholipids (cardiolipin or phosphatidic acid) in the newly formed membranes. Taking into account the available examples of membrane proliferation upon protein overproduction, together with the latest biochemical, biophysical and structural data, we explore the relationship between protein synthesis and membrane biogenesis.
Abstract from Microb (1) Cell Fact 2020 Sep 4;19(1):176. doi: 10.1186/s12934-020-01433-x.
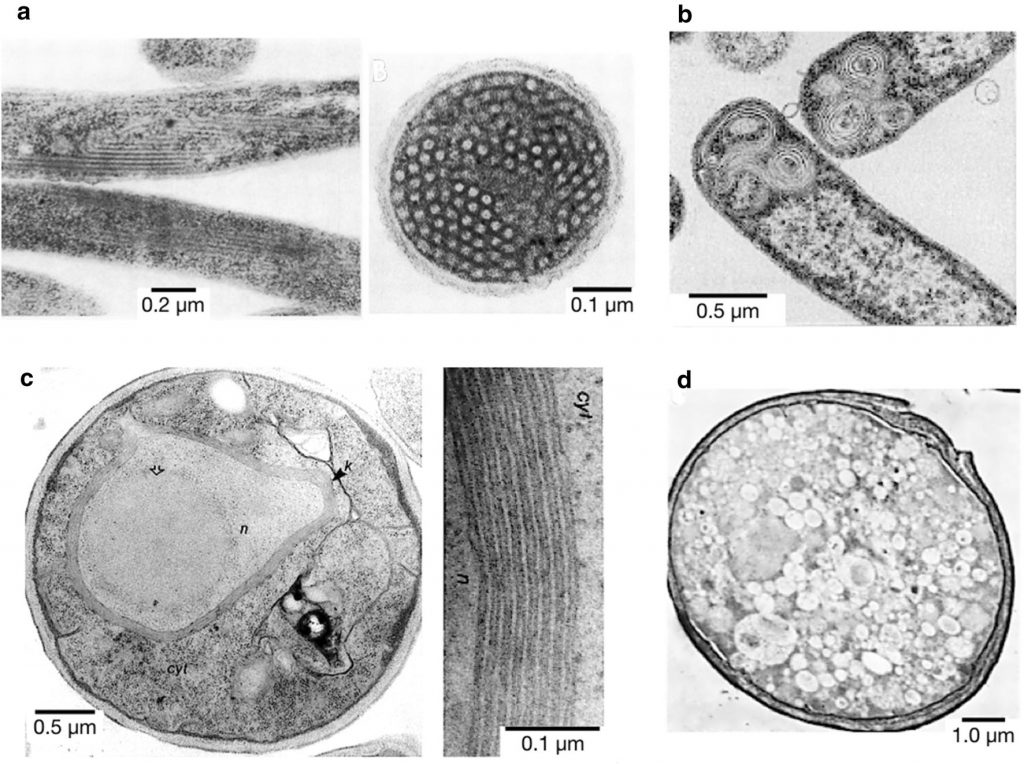
Negative-staining TEM pictures of some examples of inner membrane proliferation upon membrane protein overproduction. a Longitudinal (left) and transversal (right) sections of E. coli inner membrane tubules after fumarate reductase overproduction. b Onion-like vesicles formed upon overproduction of protein 3A of Foot and Mouth Disease Virus (FMDV). c S. cerevisiae cell with the cytosol (cyt), nucleus (n) and the stacked membranes “Karmellae” (k) around the nucleus (n) (left) and detail of those membranous structures (right) after 3-Hydroxy-3-methylglutaryl-CoA reductase (Hmg-CoA) overproduction. d Vesicles formed in S. cerevisiae upon overproduction of poliovirus protein 2BC.See (1) more details.
Our work
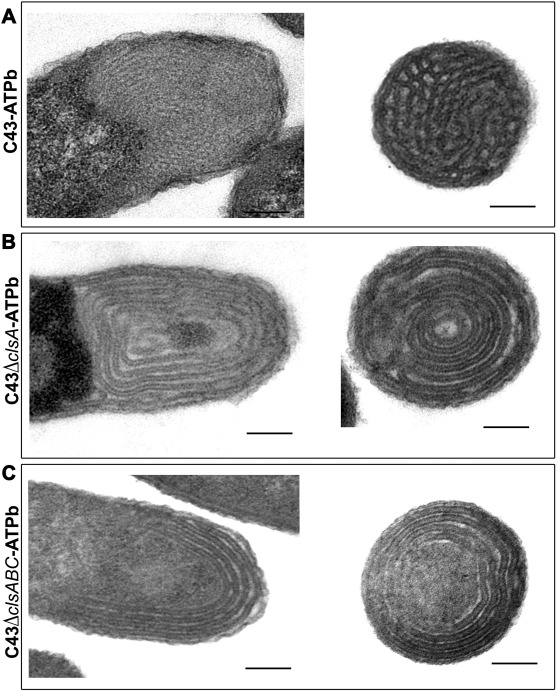
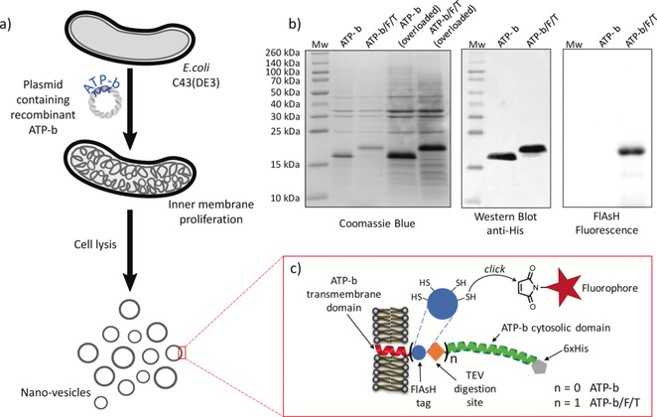
We work on the “AtpF model”. The overproduction of the b subunit of the E. coli FoF1-ATP synthase, AtpF, induces massive intracellular membrane (ICM) proliferation in C43(DE3) host (2, 3) Figure 2, and preliminary results indicate that the expression of AtpF in the chloroplast of C reinhardtii might also shape its thylakoids membrane organization. In the last years we have shown that cardiolipin play an essential role in the structuration of membranes in non-lamellar structures (3)(Figure 3). Beyond fundamental aspects of membrane biogenesis in E. coli, we have the long-term project, the “Syborg project”, to create a synthetic bacterial membrane organelle in bacteria. Applications include the in vivo structural analysis of membrane proteins, hydrophobic compounds storage, controlled encapsulation and delivery of molecules, or nano-bioreactors. For the later application we work in close collaboration with the team of Christophe Tribet (ENS chemistry department, Paris). A first step has been achieved by the conversion of the “AtpF model” into a microbial platform for high-yield production of lipidic nanovesicles with clickable thiol moieties in their outer corona. These nanovesicles show low size dispersity, are decorated with a dense, perfectly oriented, and customizable corona of transmembrane polypeptides enabling encapsulation of soluble proteins into the nanovesicles Figure 4 (4).
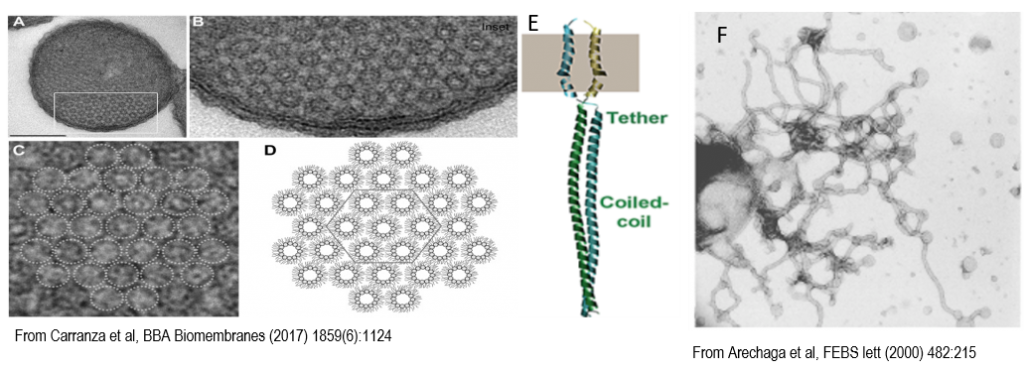
Figure 2: Nonlamellar morphology of ICMs. A, Orthogonal view of thin sections of E. coli C43(DE3) cells over-producing F-ATPase subunit b. Scale bar, 0.25 μM. B, Magnified view of the boxed area marked in A. C, Magnification of B, dotted circles indicate ICMs hexagonal structures. E, Structure of the AtpF dimer. F, Isolated membranes from ATPF overexpressing cells.
Figure 4: a) Schematic representation of the preparation of lipid:protein nanovesicles, b) SDS-PAGE analysis of isolated vesicles, and c) cartoon of ATP-b and ATP-b/F/T recombinant proteins. The FlAsH tag contains four clickable thiol sites and a TEV cleavage site enables the removal of the ATP-b cytosolic domain. From (4)
Fundings
ANR grants
- Gencaps 2017-2020 : https://anr.fr/Projet-ANR-17-CE09-0007
- FLiPosome: 2020-2024
In collaboration with
(Instituto de Biomedicina y Biotecnología de Cantabria, IBBTEC, SPAIN)
Bibliography
- Royes J, Biou V, Dautin N, Tribet C, Miroux B. 2020. Inducible intracellular membranes: molecular aspects and emerging applications. Microb Cell Factories 19:176.
- Arechaga I, Miroux B, Karrasch S, Huijbregts R, de Kruijff B, Runswick MJ, Walker JE. 2000. Characterisation of new intracellular membranes in Escherichia coli accompanying large scale over-production of the b subunit of F(1)F(o) ATP synthase. FEBS Lett 482:215–219.
- Carranza G, Angius F, Ilioaia O, Solgadi A, Miroux B, Arechaga I. 2017. Cardiolipin plays an essential role in the formation of intracellular membranes in Escherichia coli. Biochim Biophys Acta Biomembr 1859:1124–1132.
- Royes J, Ilioaia O, Lubart Q, Angius F, Dubacheva GV, Bally M, Miroux B, Tribet C. 2019. Bacteria-Based Production of Thiol-Clickable, Genetically Encoded Lipid Nanovesicles. Angew Chem Int Ed Engl 58:7395–7399.