
The hub of my research has been and is “Membrane Proteins” (MPs), from a functional point of view (cyt b6f and C. reinhardtii) and overproduction, which remains one of the major bottlenecks in structural genomics. My work is centred on two major lines. On one side, I will participate at B. Miroux projects on the approaches for the large-scale production of membrane proteins (MPs) using mutant E. coli as host. On the other side, I will exploit my expertise on the green alga C. reinhardtii and utilise this organism for two main purposes: i) to make use of C. reinhardtii chloroplast as a green factory for MPs overproduction, ii) to elucidate the signal transduction in State Transitions.
Eukaryotic membrane proteins production in E. coli
Integral membrane proteins (MP) are difficult to produce, hampering their structural and functional studies. The most versatile, and most widely used, host in structural biology of MPs, Escherichia coli, accounts for 200 of the 440 unique X-ray and NMR MP structures found in the Protein Data Bank (PDB). Several bacterial hosts as C41(DE3) and C43(DE3) (Miroux & Walker, 1999), display a reduced basal level of T7RNApol and this increases the induction fold of the expression of the target gene. Both strains have had an important impact in the structural Biology of MP (28% of non E.coli membrane protein structures, 50% of -helical MP structures (Hattab et al. 2015)) and it has been proposed that a reduced basal level of expression of the target gene should facilitate the expression of different MPs. This finding should scope out the isolation of new mutants for large amount of MPs production.
For more information on this project see Bruno Miroux’ page.
Around Chlamydomonas reinhardtii…
-
Green unicellular alga
7-8 µm, generation time 10h, microbiology techniques
-
Facultatif phototrophe (acetate):
* maintain of photosynthetic mutants * in vivo radioactive precursors incorporation
-
Accessible to genetic transformation of nucleus (aleatory recombination) and chloroplast (homologous recombination)©
-
Sexual cycle: classical genetic
-
In vivo spectroscopy/fluorescence
I am a molecular geneticist who has developed in the last 20 years a toolbox of synthetic biology techniques such as chloroplast overexpression of proteins of interest, genetics and transformation techniques. I worked out my expertise in the model organism Chlamydomonas reinhardtii, which has been for years considered the photosynthetic alter ego of yeast (Rochaix, 1995).
C. reinhardtii chloroplast as a green factory for MPs overproduction: Tuning cyanobacteria and chloroplasts for membrane proteins overexpression
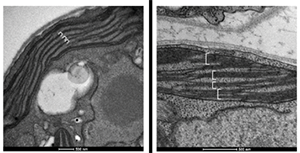
Trasmission electron microscopy of wt (A) and ATPase b subunit expressing (B) chloroplasts. Image suggest that in B the expression of subB alters the thylakoid organisation
We plan to overexpress in parallel a palette of MPs including ATPase b subunit to test its impact on membrane proliferation, Bacteriorhodopsin (BR) as model protein and uncoupling proteins (UCP) because they are difficult to produce in other expression systems.
Comparing both expression systems should gave some hints on codon usage, gene sequence, and membrane dynamic. This project will hopefully contribute to elucidate basic molecular mechanism of MPs production in the two organisms, which represent keystones in photosynthetic organisms evolution. In addition it will be intriguing to evaluate the degree of tolerance of thylakoid membranes to the “intrusion” of heterologous proteins.
Chloroplast synthetic biology for photosynthesis, metabolism and cell signaling
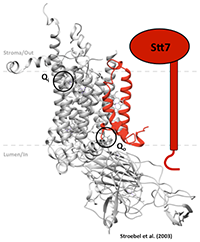
How is the redox signal transmitted across the membrane, from the Qo site to the kinase domain ?
Photosynthetic productivity is the final frontier for improving biomass production and one way to reach the goal is exploring ways of redesigning photosynthesis (Ort et al 2015).
For such an ambitious undertaking, an integrated approach is essential, starting from the molecular level and analyzing the effects of changes at increasingly organized levels. To prove the feasibility of this approach, we have chosen to target the cytochrome b6f, which represents a limiting step for photosynthesis under non-limiting CO2 concentrations. The cyt b6f is central in the regulation of both electron and proton transfer, it is also a key regulatory point for photo-protective mechanisms such as state transitions (Dumas et al 2016, 2017, 2018; Alric et al 2015; Zito et al, 1999; Zito et al 2002; de Lacroix et al 2007).
The aim of this approach is to consolidate the information on the connection among the receptor of the signal (Subunit IV of cyt b6f complex in red in the figure), the source of the signal (Plastoquinone binding site Qo in Subunit b6) and the target of the signal (STT7 kinase) using a random mutagenesis technique. Chlamydomonas reinhardtii is our organism of choice, due to ease of transformation and genetic manipulation, we have already obtained solid evidences that cytochrome b6f complex can be randomly mutagenized and reassembled de novo, a first step towards the directed evolution of chloroplast enzymes.
References
- Alric J. 2015 The plastoquinone pool, poised for cyclic electron flow? Front Plant Sci. 6:540.
- Dumas L, Chazaux M, Peltier G, Johnson X, Alric J. (2016) Cytochrome b 6 f function and localization, phosphorylation state of thylakoid membrane proteins and consequences on cyclic electron flow. Photosynth Res. 129:307-320.
- Dumas L, Zito F, Blangy S, Auroy P, Johnson X, Peltier G, Alric J (2017) A stromal region of cytochrome b6f subunit IV is involved in the activation of the Stt7 kinase in Chlamydomonas PNAS 114:12063-12068.
- Dumas L, Zito F, Auroy P, Johnson X, Peltier G, Alric J (2018) Random mutagenesis of a Chlamydomonas chloroplast gene by error-prone PCR Plant Physiol., 77: 465-475
- Hattab, G, Warschawski D.E., Moncoq K, and Miroux B. (2015). «Escherichia coli as host for membrane protein structure determination: a global analysis». Scientific Reports 5 . doi:10.1038/srep12097.
- de Lacroix de Lavalette A, Finazzi G and Zito F. (2008) b6f-Associated chlorophyll: structural and dynamic contribution to the different cytochrome functions. Biochemistry. 47: 5259-5265.
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP, Moore TA, Moroney J, Niyogi KK, Parry MA, Peralta-Yahya PP, Prince RC, Redding KE, Spalding MH, van Wijk KJ, Vermaas WF, von Caemmerer S, Weber AP, Yeates TO, Yuan JS, Zhu XG. (2015) Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc Natl Acad Sci U S A 14;112:8529-8536.
- Rochaix JD. (1995) Chlamydomonas reinhardtii as the photosynthetic yeast Annu Rev Genet. 29:209-230.
- Walker, J., and Miroux, B. (1999). Selection of Escherichia coli Hosts That Are Optimized for the Overexpression of Proteins. In Manual of Industrial Microbiology and Biotechnology, 2nd Edition (MIMB2)., (A.L. Demain and J.E. Davies)
- Zito F., Finazzi G., Delosme R., Nitschke W., Picot D. and Wollman F.-A. (1999) The Qo site of cytochrome b6f complexes controls the activation of the LHCII-kinase. EMBO J. 18 : 2961-2969
- Zito F, Vinh J., Popot J.L. and Finazzi G. (2002) Chimeric fusions of subunits IV and PetL in the b6 f complex of Chlamydomonas reinhardtii. Structural implications and Consequences on State Transitions. J. Biol. Chem. 277 : 12446-12455.