Amphipols: Amphipathic polymers that represent a detergent alternative to stabilize membrane proteins in aqueous solutions

Manuela
Zoonens
Research Director

Korantin
le Mouël
pHD STUDENT
Introduction
Studying the structure and function of membrane proteins (MPs) remains a major challenge due to the need to extract MPs from the lipid membrane, which both forms their native environment, and strongly influences their structure and function. Solubilization of biological membranes is conventionally achieved using detergents, which, in a solution of detergent-solubilized MP, are in a constant equilibrium between monomers, micelles, and the surfactant layer that covers the transmembrane region of the protein and makes it hydrophilic. To prevent MPs from aggregating, detergent must be handled above the cmc of the detergent, i.e. in the presence of free micelles. The latter act as a sink for hydrophobic and amphipathic molecules, e.g. lipids, which is a major cause of MP inactivation. This could in principle be alleviated by designing molecules with such a high affinity for the transmembrane surface of MPs that they would never dissociate. Thus, traces of free surfactant would suffice to keep the protein soluble. This concept led to the creation of ‘amphipols’ (APols), namely short amphipathic polymers that are able to keep individual MPs soluble under the form of small hydrophilic complexes without the need of adding free surfactant in buffer1.
1. APol A8-35
The prototypical APol A8-35, which is a poly(acrylic acid) (PAA) polymer randomly modified with octylamine and isopropylamine side chains is the APol of reference (Fig. 1) . The average number of acrylate units is ~35 and the average molecular mass of A8-35 is 4.3 kDa. Up to date, this polymer is the most used for determining the high-resolution structure of MPs by single-particle cryo-electron microscopy2.
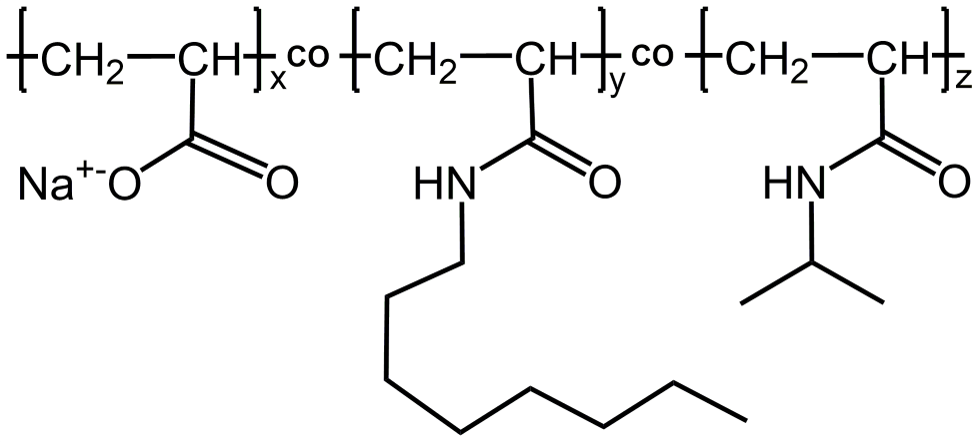
Figure 1. Chemical structure of APol A8-35; x ~ 0.35, y ~ 0.25, and z ~ 0.4 (from ref. (3)).
2. Functionalized A8-35
In addition to abolish the addition of free surfactant in buffer, another advantage of APols derives from the fact that grafting a small percentage of a tag or label onto the polymer does not affect its solution properties, while functionalizing it for specific purposes4.
A8-35 functionalized with the fluorescent probe NBD (FAPol) allowed FRET experiments to be performed5. These experiments showed that extensive dilution of MP/FAPol particles into an APol-free medium does not entail any detectable desorption of FAPols bound to the protein, even after extended periods of time (hours-days) (Fig. 2A). On the other hand, MP-bound FAPol readily exchanges for other surfactants, be they detergent or unlabelled APol (Fig. 2B).
Using functionalized APols results in functionalizing the MP of interest without the need for any chemical nor genetic modification. Many applications of such a mild labeling can be performed. For instance, functionalized A8-35 carrying affinity tags could be used to immobilized MPs onto a solid support6–8 (Fig. 3A) or label a MP for structural studies9 (Fig. 3B), whereas those carrying adjuvant can be used for vaccine applications10,11. Finally, given that APols are miscible, they offer also an interesting mode of adding several functionalities at a time to a MP by simply trapping it in a mixture of modified APols12.
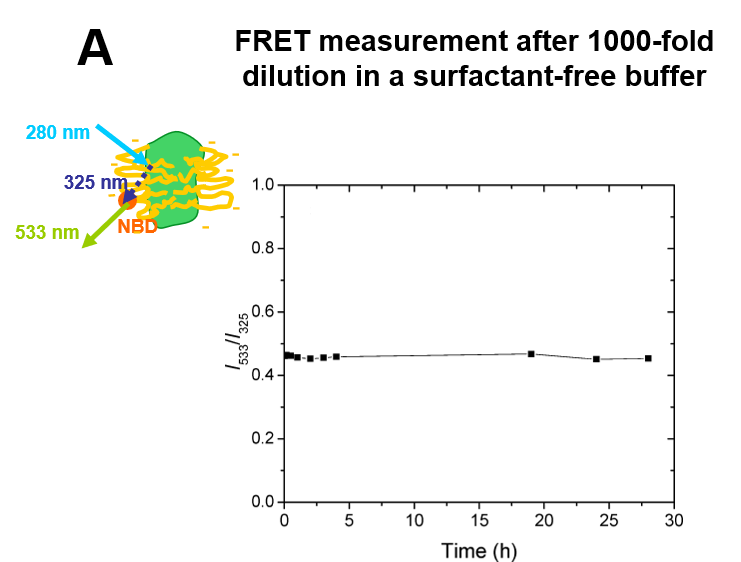
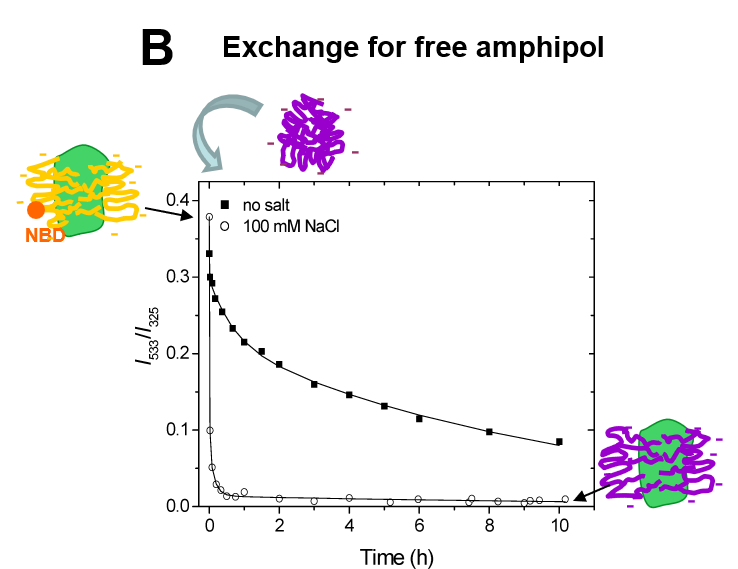
Figure 2. Dynamics of the APol layer surrounding the transmembrane domain of a MP. A. Stability of the fluorescence of tOmpA/FAPol complexes after 1000 × dilution into APol-free buffer, indicating the absence of dissociation. B. Kinetics of exchange of tOmpA-bound FAPol for unlabelled APol after supplementation of a tOmpA/FAPol sample with a 50 × excess of unlabelled APol: provided the repulsive electrostatic interactions between MP-bound and free APols are screened, the exchange is quite rapid (minutes) (from ref. (5)).
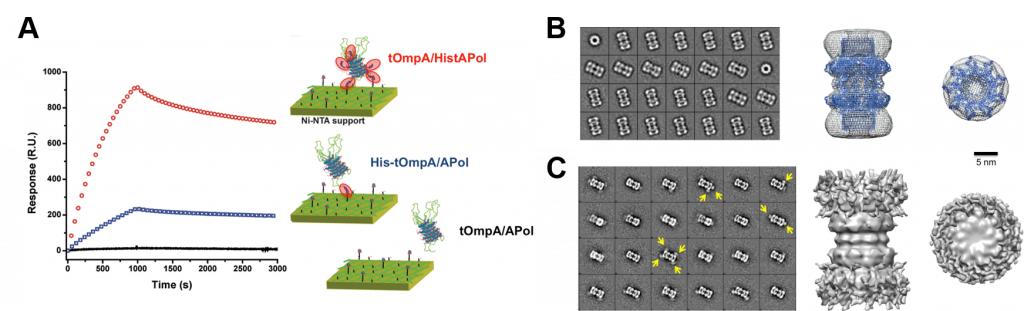
Figure 3. Some applications of functionalized APols. (A) SPR sensorgrams of different tOmpA/APol complexes injected on a Ni:NTA surface (right) and illustration of the binding interaction between the complexes and the support (left). The signal observed for tOmpA/HistAPol complexes (red curve) is compared to that measured for tOmpA with a His-tag fused at the N-terminus of the protein (blue curve) or without any tag (black curve), both versions of tOmpA trapped in plain A8-35 (from ref.(8)). (B, C) Negative stain-EM of CsgG trapped in BAPol (B) without or (C) with monovalent streptavidin (mSA) labeling. Left, representative 2D class averages of (B) CsgG/BAPol and (C) CsgG/BAPol/mSA. Yellow arrows indicate the position of mSA bound to the complex. Right, 3D reconstruction of the complexes from NS-EM images, side views and top views of the particles. The 3D models were obtained after imposing a D9 symmetry during the reconstitution process to obtain a good-quality model of CsgG, resulting in an artificial symmetrization of the mSA. The crystal structure of CsgG dimer (PDB accession code: 4uv3) was fitted into the 3D model obtained by NS-EM of CsgG/BAPol complexes. The β-barrels forming the transmembrane domains are found at the extremities of the particle and match the distribution of streptavidin (from ref.(9)).
3. CyclAPols: a new generation of APols
A8-35 is usually used subsequent to MP purification in detergent13 because its capacity at extracting MPs highly varies from one MP and from one biological membrane to another. Therefore, a new generation of APols has been designed, called CyclAPols, which differ from A8-35 mainly by the replacement of linear octyl chains for hydrophobic, saturated cycles14. This change markedly improves the polymer ability to extract MPs from biological membranes, regardless to the membrane and the protein of interest, while preserving the native structure of the target MP (Fig. 4). In addition, the CyclAPols are UV compatible in contrast to the styrene maleic acid copolymers (SMAs).
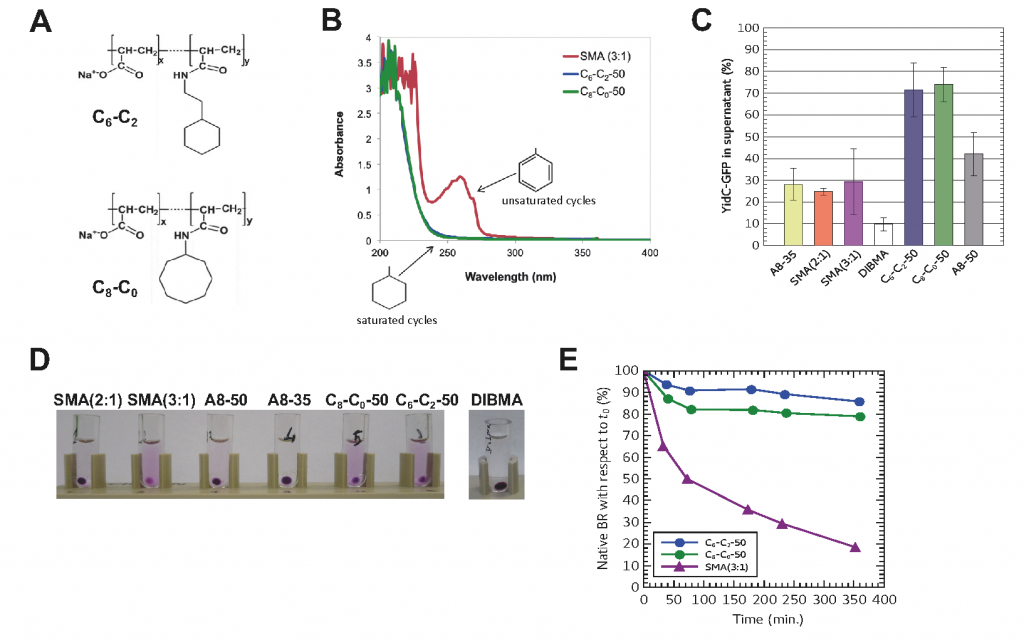
Figure 4: The CyclAPols combine the extraction efficiency of SMAs with the stabilization afforted to MPs by classical APols such as A8-35. (A) Chemical structures of two CyclAPols (C6-C2-50 and C8-C0-50). (B) UV-visible spectra of CyclAPols and SMA (3:1) solubilized at 1 mg/mL in deionized water. (C) Solubilization of YidC-GFP from the plasma membrane of E. coli. In each condition, the total MP/polymer mass ratio was 1:1. Buffer was 20 mM Tris-HCl pH 8.0, 150 mM NaCl. After 1 h incubation at 4°C, and centrifugation (30 min at 100,000 × g), the supernatant and pellet of each sample were analyzed by SDS-PAGE. The fluorescence intensity of the band corresponding to YidC-GFP in the supernatant was quantitified by imageJ. (D) Solubilization by co-polymers of bacteriorhodopsin (BR) from the DMPC-fused purple membrane of Halobacterium salinarum. Photograph of samples after an overnight incubation at 25°C followed by an ultracentrifugation (20 min. at 200.000 × g). (E) Comparison of the thermostability of BR extracted with either CyclAPols or SMA. The samples were incubated at 50°C for 6 h and their UV-visible spectrum recorded each hour. The absorbance values at 554 nm were plotted as function of the incubation time (from ref.(14)).
For more information about APols and their applications, see http://www.ibpc.fr/amphipol
For a complete list of references related to APols see: http://www.ibpc.fr/amphipol/references/references.html