
Philippe
Delepelaire
Research Director

Nathalie
Dautin
RESEARCHER

Annick
Paquelin
Technician

Valérie
Biou
Researcher

Martin
Picard
Research Director

Xiu
Tian
pHD STUDENT

Danielle
Castro
Post-doc

Caine
Tran
Engineer
Heme uptake and porphyrin homeostasis in Gram-negative bacteria: from function to structure and vice-versa
Iron is an essential component for most living organisms. Heme (iron protoporphyrin IX) is the major iron source for commensal and pathogenic bacteria that develop inside mammals as heme contained in hemoglobin represents more than 50% of all body iron. Bacteria have developed many systems to take up this molecule, that they most often use as an iron source (requiring heme iron extraction), or as a heme source.
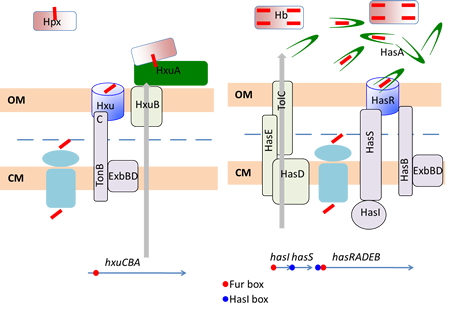
To do so, Gram-negative bacteria use the so-called TonB-Dependent Transporters (TBDT’s), found in their outer membrane, together with the TonB complex from the cytoplasmic membrane. Heme specific TBDT’s bind heme (they might also bind hemoproteins) and internalize it into the periplasm. This is an energy-requiring process powered by the proton motive force (pmf) and recruiting of the TonB complex.
Our work
Our aim is to understand how these macromolecular assemblages work, from the molecular level to their integration into the bacterial cell physiology. Two models are studied, Has (heme acquisition system) from Serratia marcescens and Hxu (Hemopexin utilization) from Haemophilus influenzae. To this end we use molecular genetics, biochemistry and we collaborate with structural biologists and biophysicists.
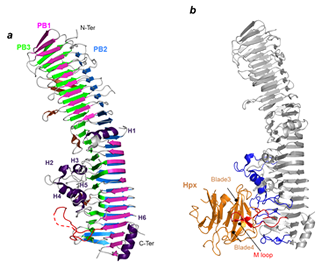
Right: Structure of the HxuA protein alone (left) and in complex with the N-terminal domain of hemopexin (right).
Bibliography
- S. Krieg, F. Huché, K.Diederichs, N. Izadi-Pruneyre, A. Lecroisey, C Wandersman, P. Delepelaire and W. Welte Heme uptake across the outer membrane as revealed by crystal structures of the receptor-hemophore complex. Proc Natl Acad Sci U S A. 106: 1045-1050.(2009)(Pubmed)
- S. Zambolin, B. Clantin, M. Chami, S. Hoos, A. Haouz,V. Villeret and P. Delepelaire Structural basis for haem piracy from host haemopexin by Haemophilus influenzae. Nat Commun. 7:11590.(2016) (Pubmed)