Bruno
Miroux
Research Director

Françoise
Bonneté
RESEARCHER

Karine Moncoq
UPC MCU

Sandrine
Masscheleyn
Enginneer

Alexandre
Pozza
Research Engineer

Rebecca Moussa
pHD STUDENT

François
Gellé
pHD STUDENT
The Uncoupling Protein 1 (UCP1)
Context:
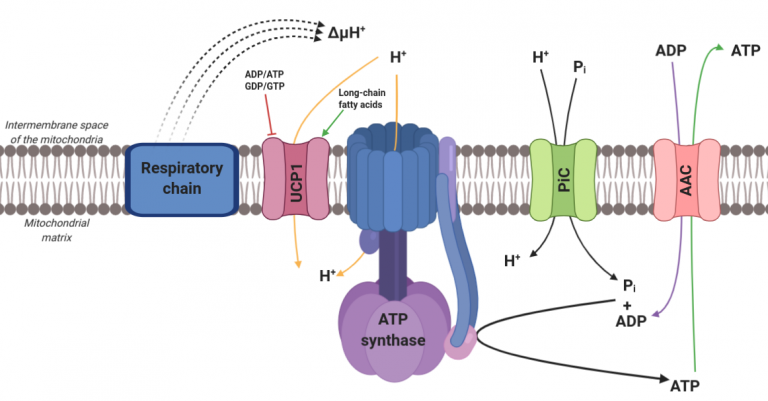
In homeotherm, regulation of metabolism activity is a way to control body temperature. In mammals, a specific tissue is dedicated to heat production upon cold exposure : the brown adipose tissue (BAT). BAT mitochondria express large amount of Uncoupling protein 1 (UCP1), which enables a proton leak from the intermembrane space to the mitochondrial matrix. Consequently, the proton permeability induced by UCP1 activity fully uncouples respiration from ATP synthesis and dissipates energy from oxidative catabolism as heat (Figure 1).
Figure 1 : Role of UCP1 in the brown adipose tissue mitochondria.
ATP synthase uses the proton gradient, as an electrochemical force, to produce ATP from ADP and free phosphate, both coming from outside the matrix by specific carriers : ADP/ATP Carrier (AAC) and Phosphate Carrier (PiC).
UCP1 bypasses the coupling between proton gradient production (electron transport chain) and use (ATP synthase). The uncoupling induced by UCP1, combined with the lack of ATP enables the electron transport chain, thus the catabolism upstream, to be faster.
Despite being identified nearly 50 years ago by Daniel Riquier [1], UCP1 structure and mechanism of proton transport remain elusive. It is part of a large protein family, the mitochondrial carriers [2], the most known being ADP/ATP carrier (AAC). Recently described structures [3,4] (PDB: 1OKC and 6GCI) have confirmed the predicted topology of this family members: 3-order pseudo-symmetry, with three homologous repeats (approximately 100 amino acids each, structured in two transmembrane alpha-helices).
Previous works have shown UCP1 is stimulated by long-chain fatty acids, and repressed by purine nucleotides [5]. However, activation and inhibition mechanisms as well as binding sites of those molecules are mainly unknown and addressing those questions represents one of the most promising leads to decipher relationship between UCP1 structure and function.
Previous works have shown UCP1 is stimulated by long-chain fatty acids, and repressed by purine nucleotides [5]. However, activation and inhibition mechanisms as well as binding sites of those molecules are mainly unknown and addressing those questions represents one of the most promising leads to decipher relationship between UCP1 structure and function.
Our approaches
Mitochondrial carriers are fragile alpha-helical proteins that are difficult to handle in presence of detergent. We have shown in the past that structural NMR models obtained in dodecylphosphocholin detergent micelles (DPC) are unstable and highly permeable to water molecules in molecular dynamic simulations and that DPC irreversibly inhibit UCP1 function [6,7]. More recently, we found that amino-acids identified by NMR titration in DPC as crucial for UCP1 activation by long chain free fatty acid are without effect in a physiological mitochondrial context [8].
Our approach combine ligand chemistry with protein refolding and functional assays on yeast mitochondria to identify key residues involved in the regulation of UCP1 activity. Long term projects include a dynamic atomic description of UCP1 in presence of regulating ligands.
Figure 2 : Graphical abstract from [8].
In collaboration with
We have a long-standing collaboration with Fréderic Bouillaud, Institut Cochin, Paris, and two modelling groups François Dehez & Chris Chipot, Nancy and Jérome Henin & Antoine Gagelin, at the Laboratory of Theorithical Biochemistry at IBPC.
Fréderic Bouillaud
(Institut Cochin, Paris)
Jérome Henin & Antoine Gagelin
(Laboratory of Theorithical Biochemistry, IBPC, Paris)
Open positions
UCP1 team is always seeking for talented young researchers, either from Bachelor or Masters degree, with a possible extension into a PhD. Any motivated candidate is encouraged to contact us to discuss about their project and opportunities in the group or the lab.
Bibliography
[1] Ricquier, D. UCP1, the Mitochondrial Uncoupling Protein of Brown Adipocyte: A Personal Contribution and a Historical Perspective. Biochimie 2017, 134, 3–8. https://doi.org/10.1016/j.biochi.2016.10.018.
[2] Robinson, A. J.; Overy, C.; Kunji, E. R. S. The Mechanism of Transport by Mitochondrial Carriers Based on Analysis of Symmetry. Proc. Natl. Acad. Sci. U.S.A. 2008, 105 (46), 17766–17771. https://doi.org/10.1073/pnas.0809580105.
[3] Pebay-Peyroula, E.; Dahout-Gonzalez, C.; Kahn, R.; Trézéguet, V.; Lauquin, G. J.-M.; Brandolin, G. Structure of Mitochondrial ADP/ATP Carrier in Complex with Carboxyatractyloside. Nature 2003, 426 (6962), 39–44. https://doi.org/10.1038/nature02056.
[4] Ruprecht, J. J.; King, M. S.; Zögg, T.; Aleksandrova, A. A.; Pardon, E.; Crichton, P. G.; Steyaert, J.; Kunji, E. R. S. The Molecular Mechanism of Transport by the Mitochondrial ADP/ATP Carrier. Cell 2019, 176 (3), 435-447.e15. https://doi.org/10.1016/j.cell.2018.11.025.
[5] Rial, E.; Poustie, A.; Nicholls, D. G. Brown-Adipose-Tissue Mitochondria: The Regulation of the 32000-Mr Uncoupling Protein by Fatty Acids and Purine Nucleotides. Eur. J. Biochem. 1983, 137 (1–2), 197–203. https://doi.org/10.1111/j.1432-1033.1983.tb07815.x.
[6] Zoonens, M.; Comer, J.; Masscheleyn, S.; Pebay-Peyroula, E.; Chipot, C.; Miroux, B.; Dehez, F. Dangerous Liaisons between Detergents and Membrane Proteins. The Case of Mitochondrial Uncoupling Protein 2. J. Am. Chem. Soc. 2013, 135 (40), 15174–15182. https://doi.org/10.1021/ja407424v.
[7] Chipot, C.; Dehez, F.; Schnell, J. R.; Zitzmann, N.; Pebay-Peyroula, E.; Catoire, L. J.; Miroux, B.; Kunji, E. R. S.; Veglia, G.; Cross, T. A.; Schanda, P. Perturbations of Native Membrane Protein Structure in Alkyl Phosphocholine Detergents: A Critical Assessment of NMR and Biophysical Studies. Chem Rev 2018, 118 (7), 3559–3607. https://doi.org/10.1021/acs.chemrev.7b00570.
[8] Piel, M. S.; Masscheleyn, S.; Bouillaud, F.; Moncoq, K.; Miroux, B. Structural models of mitochondrial uncoupling proteins obtained in DPC micelles are not functionally relevant, The FEBS journal 2020, https://doi.org/10.1111/febs.15629.