Laurent J.
Catoire
Research Director

Miri
Garrigues
pHD STUDENT

Claudia
Zilian
Post-doc

Nahya
Nguyen Armengaud
M2 Student
Study of the energy landscape of membrane proteins: application to seven trans-membrane α-helix receptors
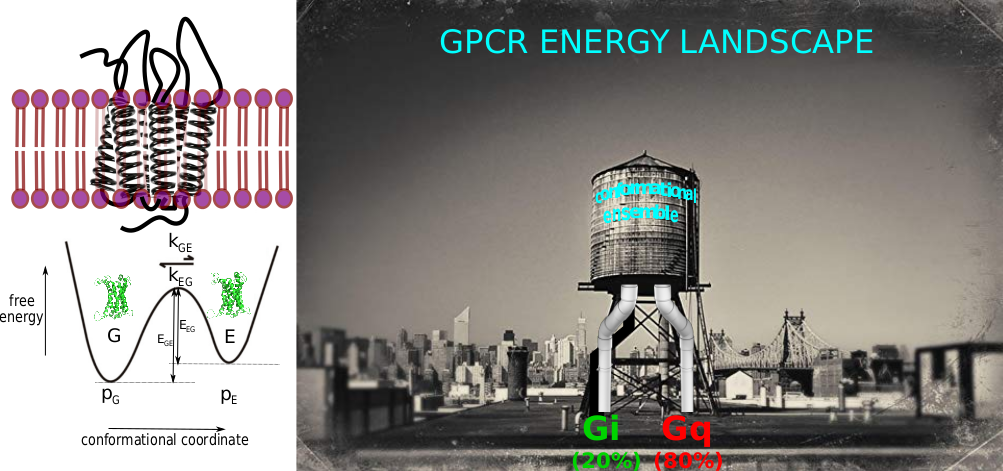
To fully characterize a biomolecule, and so to decipher its function, usually the solely atomic description of a low energy, i.e. predominantly populated and long-lived, conformational state is not enough [1]. Indeed, countless biological functions are closely linked to changes in spatial and temporal location of groups of atoms in biomolecules. In the formalism of an energy landscape, to characterized a biomolecule at a molecular level, it is essential to: i) census the number of conformations co-existing or existing along the functional pathway; ii) to get a 3D description of these structures; iii) to determine the relative probabilities of existence of these different sub-states (thermodynamics); iv) to detect and characterize chemical exchanges, i.e. the kinetic barriers separating these sub-states and associated energies of activation (kinetics).
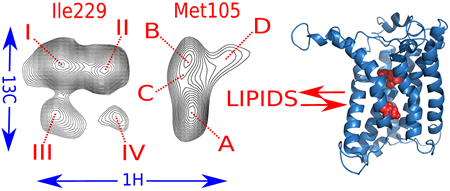
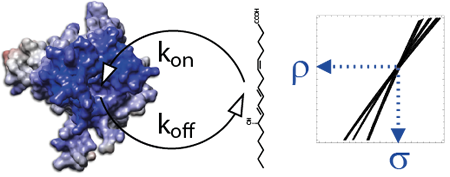
Cell membranes represent a complex and variable medium in time and space of lipids and proteins. Their physico-chemical properties are determined by lipid components which can in turn influence the biological function of membranes. Here, we used hydrostatic pressure to study the close dynamic relationships between lipids and membrane proteins. Experiments on the β–barrel OmpX and the α–helical BLT2 G Protein-Coupled Receptor in nanodiscs of different lipid compositions reveal conformational landscapes intimately linked to pressure and lipids. Pressure can modify the conformational landscape of the membrane protein per se, but also increases the gelation of lipids, both being monitored simultaneously at high atomic resolution by NMR (from [7]).
In the lab, we focus on the energy landscape of membranes proteins in lipid bilayer environments, with some recent examples that concern eukaryotic α-helical G Protein-Coupled Receptor (GPCRs) and prokaryotic β barrel proteins in nanometric lipid bilayers (nanodiscs) [2-7].
Bibliography (in blue from the lab):
- Sekhar & Kay (2013). NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc Natl Acad Sci USA, 110, 12867-74 (Pubmed)
- Casiraghi et al. (2016) Functional modulation of a GPCR conformational landscape in a lipid bilayer. J Am Chem Soc, 138, 11170-5 (Pubmed)
- Casiraghi M, Damian M, Lescop E, Banères JL, Catoire LJ. (2018) Illuminating the Energy Landscape of GPCRs: The Key Contribution of Solution-State NMR Associated with Escherichia coli as an Expression Host. Biochemistry 57, 2297-307
- Casiraghi M, Point E, Pozza A, Moncoq K, Banères JL, Catoire LJ. (2019) NMR analysis of GPCR conformational landscapes and dynamics. Mol Cell Endocrinol, 484, 69-77
- Louet M, Casiraghi M, Damian M, Costa MG, Renault P, Gomes AA, Batista PR, M’Kadmi C, Mary S, Cantel S, Denoyelle S, Ben Haj Salah K, Perahia D, Bisch PM, Fehrentz JA, Catoire LJ, Floquet N, Banères JL. (2021) Concerted conformational dynamics and water movements in the ghrelin G protein-coupled receptor. Elife 10, e63201
- Damian M, Louet M, Gomes AAS, M’Kadmi C, Denoyelle S, Cantel S, Mary S, Bisch PM, Fehrentz JA, Catoire LJ, Floquet N, Banères JL. (2021) Allosteric modulation of ghrelin receptor signaling by lipids. Nat Commun, 12, 3938
- Pozza et al. (2022) Exploration of the dynamic interplay between lipids and membrane proteins by hydrostatic pressure. Nat Commun, 13, 1780